HELP INDEX
This tool was developed for high-throughput, rational sgRNA design in CRISPR screen experiments.
Input
Species selection: Human or Mouse.
Number of sgRNAs for each gene: 1-15.
Gene IDs: could be either official gene symbol or RefSeq ID, maximum 1000.
SgRNA length: 19 or 20 nucleotides.
Also design control sgRNAs: if selected, both positive and negative control sgRNAs will be appended to the output, whose number is in proportion to the number of total sgRNA, maximum 5000.
Avoid cancer recurrent mutations sites (for homo sapiens only): if selected, sgRNAs which are overlapped with cancer recurrent mutations will be filtered out.
Add promoter/scaffold sequences to the spacer: if selected, human U6 promoter as well as spCas9 scaffold sequence will be attached to the spacers in output.
Output
sgID: a unique ID for each sgRNA within result file.
seq: sgRNA sequence.
chrom: chromosome name where the target sequence is located.
start: the start coordinate of sgRNA sequence (1-based).
end: the end coordinate of sgRNA sequence (1-based).
strand: strand information, either + or -.
efficiency: the efficiency score of sgRNA sequence (described in detail below).
conservation: the conservation score of sgRNA sequence (described in detail below).
specificity: the specificity score of sgRNA sequence (described in detail below).
offhit_nonexon: number of putative off-target loci (in terms of perfect match) which hit non-exon region within genome.
offhit_noncode: number of putative off-target loci (in terms of perfect match) which hit exon but non-coding region within genome.
offhit_code: number of putative off-target loci (in terms of perfect match) which hit coding region within genome.
Indices of sgRNA performance
Efficiency: The cleavage efficiency of an sgRNA is a major factor that determines the sensitivity of a screen experiment. Here in CRISPRscreen SSC, a computational algorithm that we previously developed [1], is applied to predict the cleavage efficiency of candidate sgRNAs. SSC takes spacer sequence together with its 3' flanking region as input, and uses Least Absolute Shrinkage and Selection Operator (LASSO) model to calculate an efficiency score for each sgRNA. CRISPRscreen will filter sgRNAs with efficiency score below zero.
Specificity: For each candidate sgRNA, firstly a specificity score was calculated according to the formula described in [2] to evaluate the overall similarity of its sequence with putative off-target genomic loci.This specificity score is ranged from 0 to 100 and the higher score shows a lower off-target effect for the spacer.
Conservation: Regions with higher conservation rates across species are more likely to be important, and they usually encode functional domains (like catalytic center for enzyme or DNA binding domain for transcriptional factor) whose knockout are more likely to disrupt gene function [3]. In CRISPRscreen, we annotated each sgRNA with an average phastCon conservation score of the corresponding target position.
SgRNA selection and ranking procedure
While a gene list is fed as input, CRISPRscreen will first retrieve all exon coordinates corresponding to the target gene, and search all sgRNA candidates that fall within these genomic features. After that, it will act in a 'filter and rescue' manner to rank all candidates and choose the top ones. For the filtering step, CRISPRscreen will filter sgRNAs that could be empirically regarded as 'bad' candidates, including sgRNAs that: 1) overlap with a SNP or mutation loci, 2) contains over 40% guanine counts ('G's), which is predicted to have higher off-target effects , and 3) are perfectly matched to putative off-target loci within the genome. The remaining ones will be ranked by a summary score, which is the a weighted summary of efficiency, specificity, conservation score as well as exon commonality score, while all the weights are dynamically defined by the CRITIC Criteria Importance Through Intercriteria Correlation (CRITIC) method [4]. For short, the purpose of CRITIC method is to determine objective weight for each criterion in multiple criteria decision problems, based on the quantification of the contrast intensity and conflicting character of evaluating criteria.
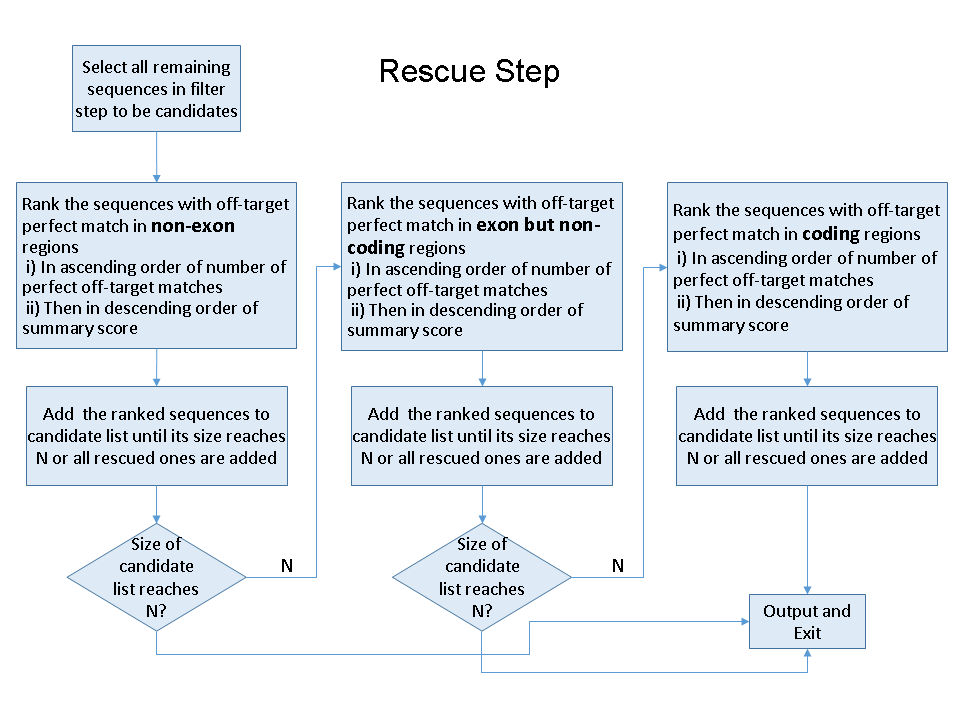
If the number of remaining sgRNAs is not enough, CRISPRscreen will use execute a 'rescue' step, trying to retrieve more possible sgRNAs. In this stage, sgRNAs with potential off-target hits will be rescued in the following order: 1) sgRNAs with non-exon off-target hits only, 2) sgRNAs with off-target hits located on non-coding elements but not coding genesregions, 3) sgRNAs with off-target hits located on coding genesregions. sgRNAs within the same category will be rescued based on their number of off-target hits, or by the summary score if two candidates have the same number of hits within the same category.
A detailed flowchart of the whole procedure is depicted in the figure below.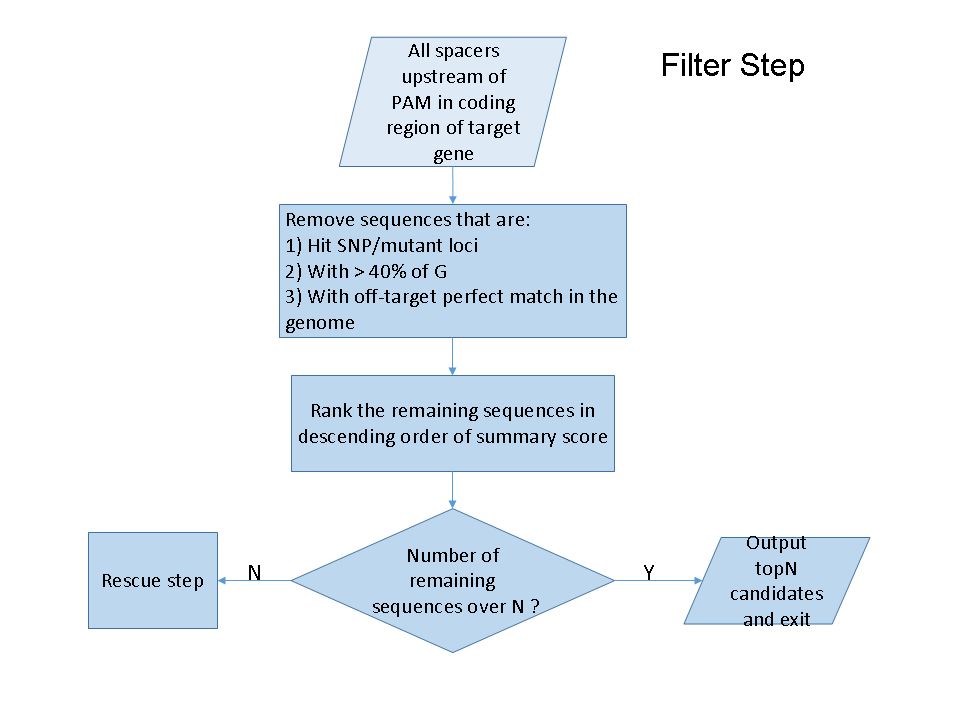
References
1. Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, et al. Sequence determinants of improved CRISPR sgRNA design. Genome research. 2015;25(8):1147-57. doi: 10.1101/gr.191452.115. PMID: 26063738.
2. Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31(9):827-32. doi: 10.1038/nbt.2647. PMID: 23873081.
3. Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome research. 2005;15(8):1034-50. doi: 10.1101/gr.3715005. PMID: 16024819.
4. Diakoulaki D, Mavrotas G, Papayannakis L. Determining objective weights in multiple criteria problems: The critic method. Computers & Operations Research. 1995;22(7):763-70. doi: http://dx.doi.org/10.1016/0305-0548(94)00059-H.
- Introduction
- Input
- Output
- Indices of sgRNA performance
- SgRNA selection and ranking procedure
- References
This tool was developed for high-throughput, rational sgRNA design in CRISPR screen experiments.
Input
Species selection: Human or Mouse.
Number of sgRNAs for each gene: 1-15.
Gene IDs: could be either official gene symbol or RefSeq ID, maximum 1000.
SgRNA length: 19 or 20 nucleotides.
Also design control sgRNAs: if selected, both positive and negative control sgRNAs will be appended to the output, whose number is in proportion to the number of total sgRNA, maximum 5000.
Avoid cancer recurrent mutations sites (for homo sapiens only): if selected, sgRNAs which are overlapped with cancer recurrent mutations will be filtered out.
Add promoter/scaffold sequences to the spacer: if selected, human U6 promoter as well as spCas9 scaffold sequence will be attached to the spacers in output.
Output
sgID: a unique ID for each sgRNA within result file.
seq: sgRNA sequence.
chrom: chromosome name where the target sequence is located.
start: the start coordinate of sgRNA sequence (1-based).
end: the end coordinate of sgRNA sequence (1-based).
strand: strand information, either + or -.
efficiency: the efficiency score of sgRNA sequence (described in detail below).
conservation: the conservation score of sgRNA sequence (described in detail below).
specificity: the specificity score of sgRNA sequence (described in detail below).
offhit_nonexon: number of putative off-target loci (in terms of perfect match) which hit non-exon region within genome.
offhit_noncode: number of putative off-target loci (in terms of perfect match) which hit exon but non-coding region within genome.
offhit_code: number of putative off-target loci (in terms of perfect match) which hit coding region within genome.
Indices of sgRNA performance
Efficiency: The cleavage efficiency of an sgRNA is a major factor that determines the sensitivity of a screen experiment. Here in CRISPRscreen SSC, a computational algorithm that we previously developed [1], is applied to predict the cleavage efficiency of candidate sgRNAs. SSC takes spacer sequence together with its 3' flanking region as input, and uses Least Absolute Shrinkage and Selection Operator (LASSO) model to calculate an efficiency score for each sgRNA. CRISPRscreen will filter sgRNAs with efficiency score below zero.
Specificity: For each candidate sgRNA, firstly a specificity score was calculated according to the formula described in [2] to evaluate the overall similarity of its sequence with putative off-target genomic loci.This specificity score is ranged from 0 to 100 and the higher score shows a lower off-target effect for the spacer.
Conservation: Regions with higher conservation rates across species are more likely to be important, and they usually encode functional domains (like catalytic center for enzyme or DNA binding domain for transcriptional factor) whose knockout are more likely to disrupt gene function [3]. In CRISPRscreen, we annotated each sgRNA with an average phastCon conservation score of the corresponding target position.
SgRNA selection and ranking procedure
While a gene list is fed as input, CRISPRscreen will first retrieve all exon coordinates corresponding to the target gene, and search all sgRNA candidates that fall within these genomic features. After that, it will act in a 'filter and rescue' manner to rank all candidates and choose the top ones. For the filtering step, CRISPRscreen will filter sgRNAs that could be empirically regarded as 'bad' candidates, including sgRNAs that: 1) overlap with a SNP or mutation loci, 2) contains over 40% guanine counts ('G's), which is predicted to have higher off-target effects , and 3) are perfectly matched to putative off-target loci within the genome. The remaining ones will be ranked by a summary score, which is the a weighted summary of efficiency, specificity, conservation score as well as exon commonality score, while all the weights are dynamically defined by the CRITIC Criteria Importance Through Intercriteria Correlation (CRITIC) method [4]. For short, the purpose of CRITIC method is to determine objective weight for each criterion in multiple criteria decision problems, based on the quantification of the contrast intensity and conflicting character of evaluating criteria.
If the number of remaining sgRNAs is not enough, CRISPRscreen will use execute a 'rescue' step, trying to retrieve more possible sgRNAs. In this stage, sgRNAs with potential off-target hits will be rescued in the following order: 1) sgRNAs with non-exon off-target hits only, 2) sgRNAs with off-target hits located on non-coding elements but not coding genesregions, 3) sgRNAs with off-target hits located on coding genesregions. sgRNAs within the same category will be rescued based on their number of off-target hits, or by the summary score if two candidates have the same number of hits within the same category.
A detailed flowchart of the whole procedure is depicted in the figure below.


References
1. Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, et al. Sequence determinants of improved CRISPR sgRNA design. Genome research. 2015;25(8):1147-57. doi: 10.1101/gr.191452.115. PMID: 26063738.
2. Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31(9):827-32. doi: 10.1038/nbt.2647. PMID: 23873081.
3. Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome research. 2005;15(8):1034-50. doi: 10.1101/gr.3715005. PMID: 16024819.
4. Diakoulaki D, Mavrotas G, Papayannakis L. Determining objective weights in multiple criteria problems: The critic method. Computers & Operations Research. 1995;22(7):763-70. doi: http://dx.doi.org/10.1016/0305-0548(94)00059-H.